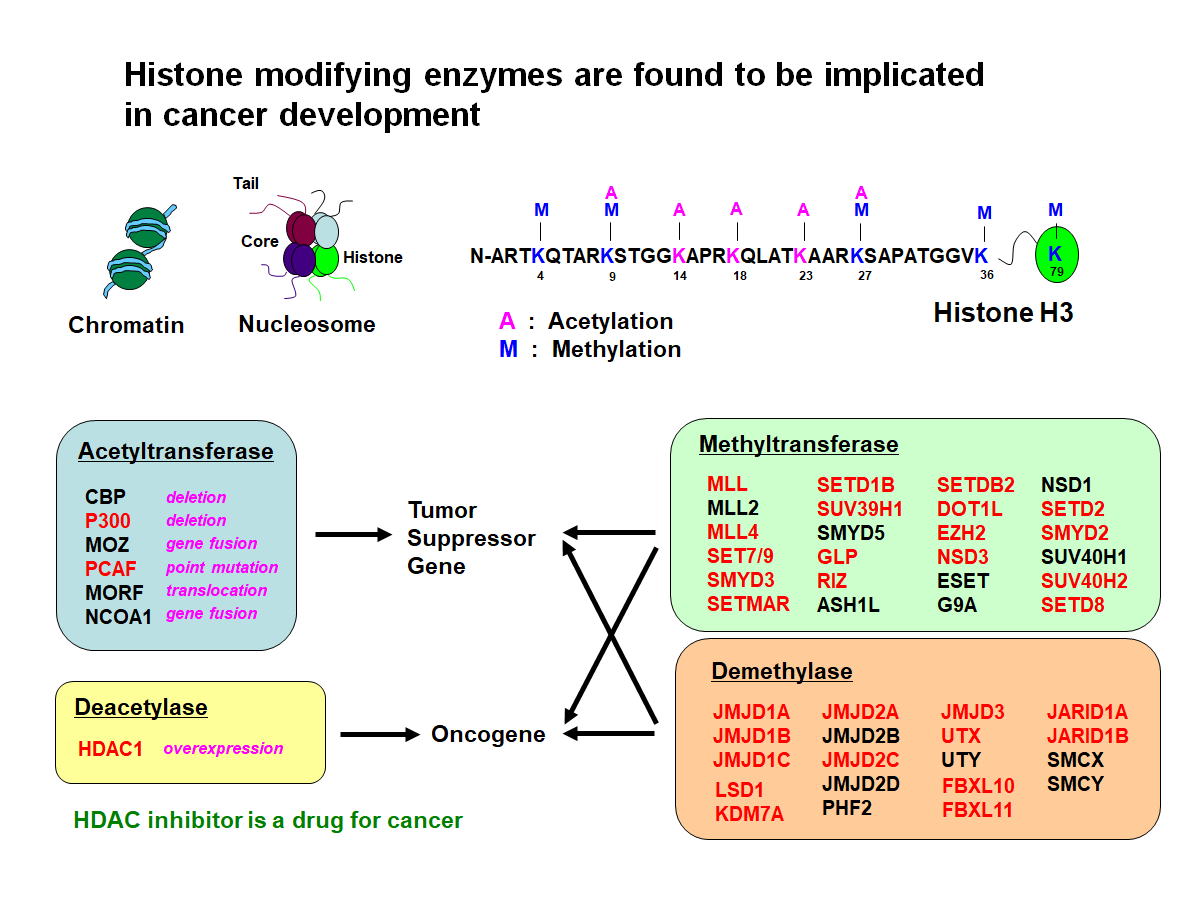
Fig.1 Numerous genes encoding histone methyltransferases and demethylases
have been identified as targets of retroviral insertional mutagenesis in
mice
Histone modifications play crucial roles in
regulating gene expression and genome function by creating global chromatin
environments. The methylation of four lysine residues (K4, K9, K27, and K36) on
the histone H3 tail is controlled by a variety of histone methyltransferases
and demethylases. Notably, many of these genes (highlighted in red) were found
to be targets of retroviral integrations, underscoring their significant roles
in oncogenesis.
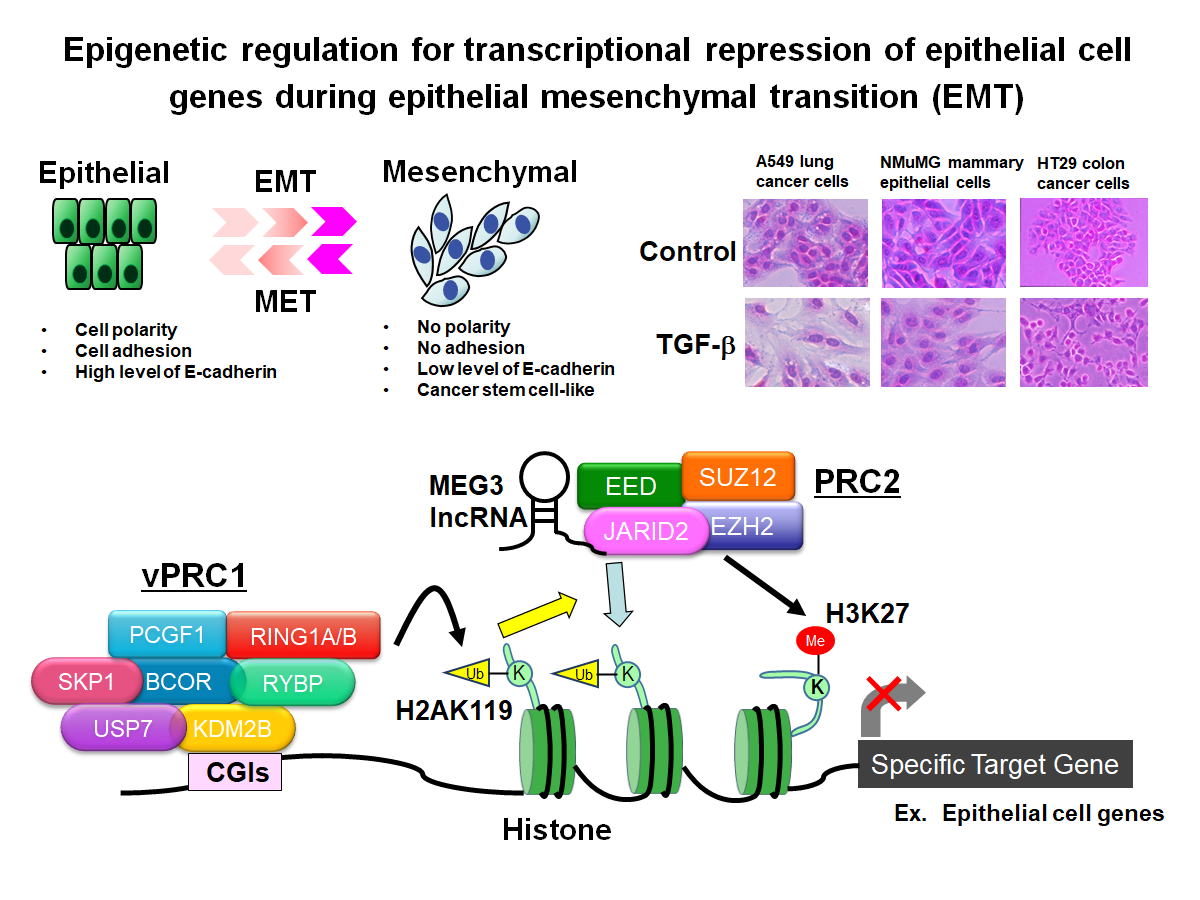
Fig.2 Epigenetic mechanisms involved in the transcriptional repression
of epithelial cell genes during epithelial mesenchymal transition (EMT)
of cancer cells
The epithelial-mesenchymal transition (EMT)
is characterized by the transformation of epithelial cells into highly motile
mesenchymal cells due to the loss of intercellular adhesion, and is regarded as
a trigger for cancer metastasis. Throughout EMT, epithelial gene
expression, including genes like E-cadherin, is repressed, while mesenchymal
genes, such as N-cadherin and Vimentin, are upregulated. Our research has
demonstrated that multiple epigenetic regulators, including the PRC2 histone
methyltransferase complex, the PRC1 histone ubiquitination enzyme complex, and
various long non-coding RNAs, play roles in the transcriptional suppression of
epithelial genes.