Organization
Division of Functional Genomics
Staff

Professor
SUZUKI, Takeshi

Assistant Professor
ISHIMURA, Akihiko
A detailed knowledge of the genes and signaling pathways mutated in cancer will be required to develop the novel target-based cancer therapeutics. However, the heterogeneity and complexity of genomic alterations in most human cancers hamper straightforward identification of cancer-causing mutations. We use the retrovirus-infected mice as model systems for identifying new cancer genes efficiently. Retroviruses induce tumors through activation of proto-oncogenes or inactivation of tumor suppressor genes as a consequence of retroviral integrations into host genome. Thus the viral integration sites provide powerful genetic tags for cancer gene identification. We are exploring the novel molecular targets for cancer treatment based on functional characterization of the cancer genes isolated by high-throughput screens using retroviral insertional mutagenesis. Once these genes are identified, we use gene knockout and transgenic mice to understand how these genes function in tumorigenesis, and to develop new animal models for human cancer. Our current projects are as follows.
- Identification of novel cancer genes using retroviral insertional mutagenesis in mice
- Involvement of histone methyltransferases and demethylases in the initiation and progression of cancer
- The role of long non-coding RNAs in malignant progression of cancer
- Relationship between RNA methyl-modifying factors and malignant progression
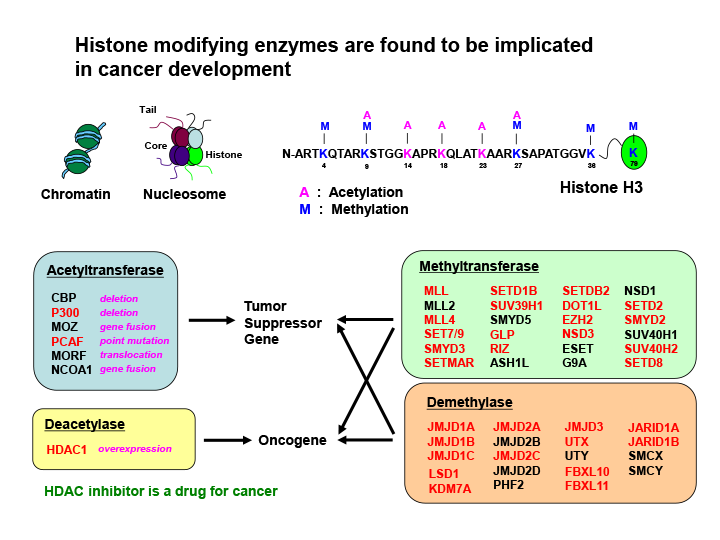
Fig.1 Most of the genes that encode histone methyltransferases and demethylases are shown to be the targets of retroviral insertional mutagenesis in mice
Histone modifications have important roles in regulating gene expression and genome function by establishing global chromatin environments. The methylation of four lysine (K) residues on the tail of histone H3 (K4, K9, K27 and K36) is regulated by a large number of histone methyltransferases and demethylases. Among them, most of the genes (shown in red) were identified as the targets of retroviral integrations, which indicates their important roles in oncogenesis.
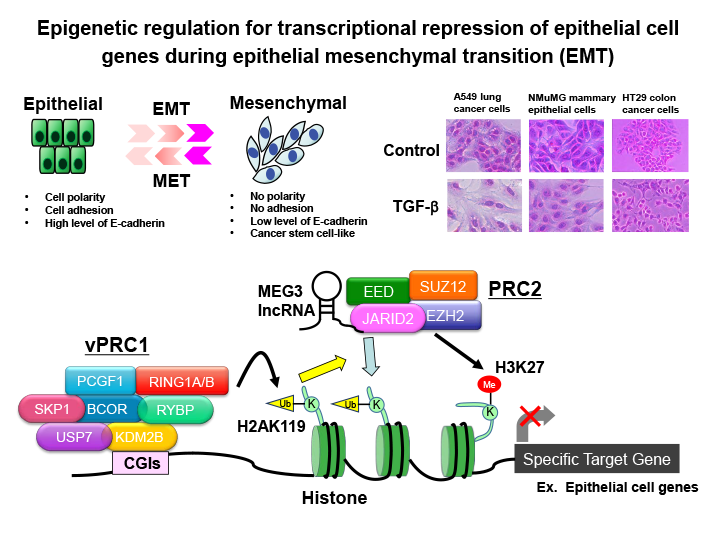
Fig.2 Epigenetic regulation for transcriptional repression of epithelial cell genes during epithelial mesenchymal transition (EMT) of cancer cells
Epithelial-mesenchymal transition (EMT), which refers to the transformation of epithelial cells into highly motile mesenchymal cells by the loss of intercellular adhesion, is considered to be a trigger for cancer metastasis. During the progression of EMT, the transcriptional suppression of epithelial genes such as E-cadherin and the upregulation of mesenchymal genes such as N-cadherin and Vimentin occur. We have shown that various epigenetic regulators such as PRC2 histone methyltransferase complex, PRC1 histone ubiquitination enzyme complex, and long non-coding RNAs are involved in the transcriptional repression of epithelial genes.

